If you have been following my article keenly, you would observe that, I have been gradually drifting from the commonly known to the less unknown here (Probably). This is aimed at making hive and @stemsocial in particular a true repository of facts and information that might not be easily gotten on the internet. Some of the information might be seen online but likely not as easy to understand as with the one you get on @Stemsocial. This is my simple long term goal and I believe it is achievable.
Today's discussion is really complex but I will as usual make it very easy to read and understand. Our discussion will bother around genes and genomics data. This is an area that is relatively virgin not until now. These data are not just gotten anyhow nor are they readable with ease rather, they are a product of complex throughput system that sequence these data from genetic materials.
Further more, today's discussion will go deeper into advances in medicine, biology, technology etc. thus, might look complicated. Regardless, I will as usual make it more friendly and easy to read. Reading this article will as usual keep you ahead when it comes to information relating to medical biotechnology - a discipline that incorporates and intersect between biology and technology. It takes advantage of the potentials a biological systems possess to produce and make advances in technology and this in turn iS dependent on different fields, e.g Agriculture, medicine, engineering etc.
A method of data extraction technique from biological sampple
In the medical world, biotechnology is the subject area that gave rise to a more detailed aspect referred to as bioinformatics. Processes involved in bioinformatics help biomedical scientist in extracting useful biological data that have changed the landscape of medical diagnosis. Useful data gotten from biological samples help in accurate diagnosis of several disease conditions that have long posed serious threat to health.
Discussing big data without starting with bioinformatics definitely will keep you the reader afloat. So let's briefly take a short trip into the world of bioinformatics, a very new and fast growing aspect of medical biotechnology that helps produce the BIG DATA which we will still be discussing at the long run.
Predicting the past, present and future through Bioinformatics
The National Center for Biotechnology Information (NCBI 2001) defined Bioinformatics as the field of science in which biology, computer science, and information technology merge into a single discipline.
Bioinformatics is more or less an interdisciplinary field whose main goal is to develop software tools and methods for the sole purpose of understanding biological data. It represents a new growing area of science that uses computational approaches to answer biological questions. Genes are the bases of inheritance in all living things and its formation is a process that is highly regulated.
Alteration in the genes produces abnormalities that results to disease conditions. The ability to detect an abnormality or a variation from the norm lies in the computational results derived from bioinformatic data.
Data in general terms simply refers to information, facts and figures collected from reliable sources for interpretation and making scientific inference. Our focus today is with regards to data whose sources are of genetic origin. Genetic data are more complex, highly voluminous and not so easy to handle unlike other data and this is one of the reasons it is referred to as Big data.
These data can be in any format and can be gotten from different sources ranging from Humans, animals and even plants etc. Data gotten at the first instance are usually crude and make no sense until they are analyzed and interpreted. This is why modern statistical tools are indispensable in data analysis. The generation of these data and their use in answering a whole lot of genetic questions especially associated with diseases is what bioinformatics aims to achieve.
The origin of these genomic data began way back in the 1950's through the ingenious works of people like James Whatson, Francis Crick, Rosaland Franklin and Maurice Wilikins who contributed to the discovery of the double helical structure of the DNA. Later on, in 1990, a very big project no one ever thought would be possible was started - proposed sequencing of the human entire genome (i.e to determine all the base pairs that is found in humans and as well extract all of them after identification).
This project was later successfully completed in the year 2003 with the efforts and collaboration of different countries and scientist all around the world. This singular ability to sequence all the genetic information from humans, a concept known as Genomics - a study of the human genome (the complete set of DNA in a living thing) has opened more doors to more findings.
Haemophilus Influenza's entire genome was sequenced (extracted) first ever in 1995 and till date, more and more organism's genome have been sequenced.

An Illumina HiSeq 2000 sequencing machine
During the 1990's, most of the processes where done manually and this limited the number of genes sequenced. By 2006, the first machine - Illumina, that was able to automatically sequence genes was produced and this drastically improved the whole process and by the year 2015, over 1000 genome project were already on. As of 2018, over 5000 genomes had already been sequenced.
Data from the human genome project has made us understand that humans have a total of about 3 billion DNA bases. Since we are able know the normal genes in humans and some living things in general, it makes it easier for us to detect and identify genetic mutations (alterations in genetic components). The possibility of knowing the normal and comparing it with the abnormal makes diagnosis of diseases more efficient, accurate and precise.
The ability to sequence all the genetic information especially in humans have paved huge way in disease diagnosis and treatment. Today, with genomic data, we can confidently predict the aetiology, prognosis (the likely outcome of a disease condition, may be positive or negative) and right individual specific treatment (precision medicine).
In general, biological data sourced or collected from genetic materials and humans to be specific are all referred to as -omics data. Now, depending on the particular structural components of the living things, or where they collected, they are named accordingly.
The commonly known ones are the as metagenomics (involves studying genetic material that is gotten from samples in the environment), proteomics (study of the protein structure and function), genomics (studying the whole complete genome), metabolomics (study of all the chemical processes that go on in the organism) and transcriptomics (studying how the organism has transcribed its DNA information to the messenger RNA, mRNA through a process known as transcription).
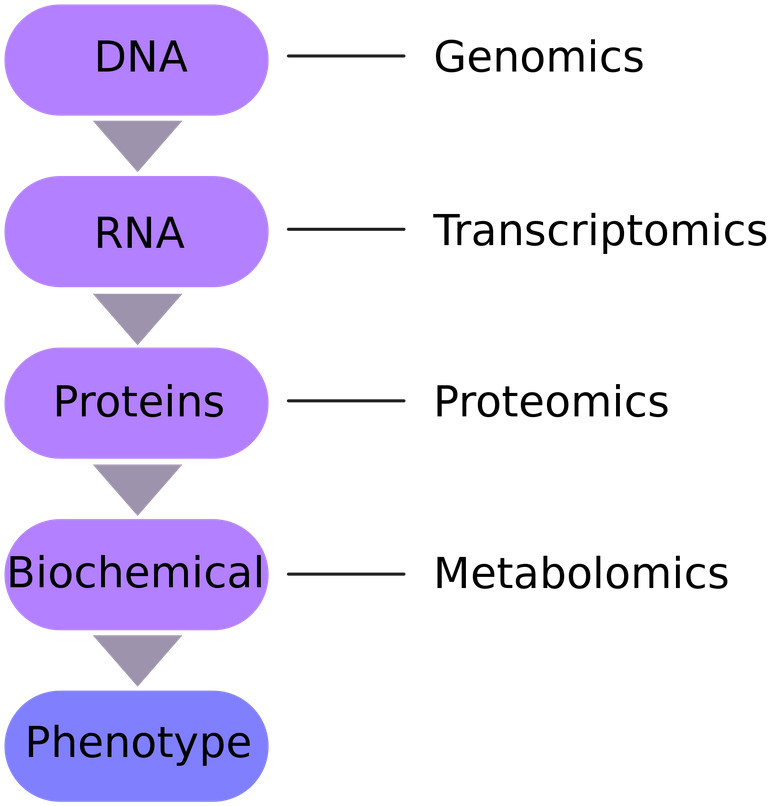
Different sources of omics data
All the data gotten from these various sources are very large and voluminous and useful in diagnosis and treatment of genetic disorders when they are analyzed.
The world of Big Data
So what are really big data? To a lay man, it could mean having plenty of data for browsing because that the commonest word used among mobile phone users when it comes to browsing and surfing the internet. To a statistician, it could mean raw information and variables to be analyzed with statistical tools. In essence, it is all correct and its meaning depends on the context in which it is applied or used.
All the data gotten from these various sources and genetic material as we have pointed out above, are really very large and voluminous thus, making it impossible for human to manually analyze. It will only take an advanced statistical tools to be able to analyze such data. Because of these, they are generally referred to as the BIG DATA.
Statistical tools are indispensable in the analysis of big data because they possess some unique and easily identifiable features. We will briefly look at some of the main key features of a big data. They are commonly known as the five V's of a big data. They include Velocity, Variety, Veracity, Volume and Value. Before any data can be tagged big data, these unique attributes are never found wanting.
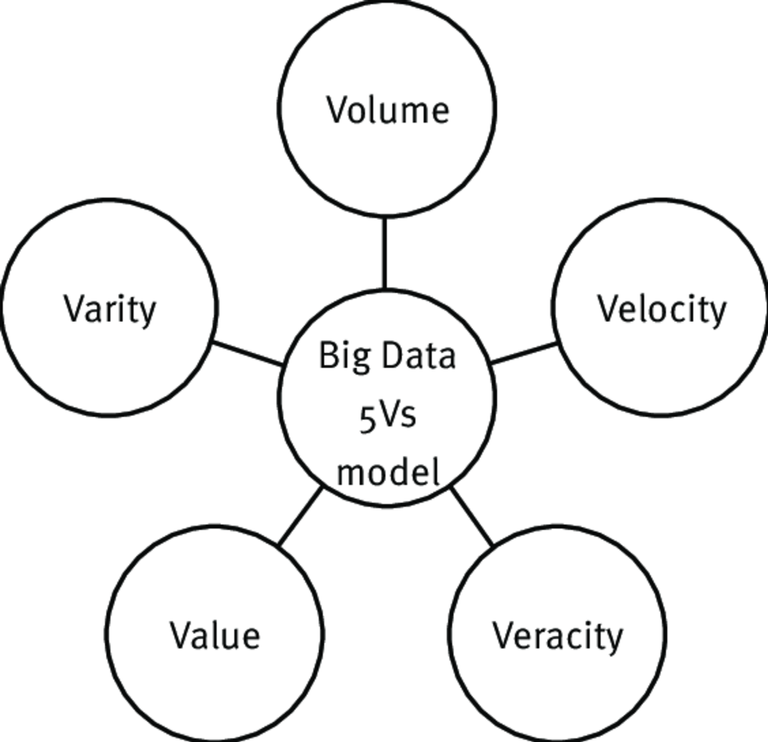
The model that currently depict big data
Genomics data can be referred to as big data because of their nature as we have seen earlier on. We will briefly look at the intricate attributes of a big data as it concerns genomics. In no particular order, we will begin first with the Velocity
Omics data are generally analyzed in modern times through high speed throughput system that systematically analyses biological data as a very fast pace. The ability to analyze these data at an ever increasing speed as they are constantly generated without having to store them in the database is remarkable.
Moving further, talking about size or volume Big data files are very voluminous and to even think of having them stored in our personal laptops will require a very large amount of disc space. For this reason, most of the big data files generated from biological sources are preferably stored in databases and subsequently made available for use by the general public. For example, platforms like Genbank, NCBI etc store data gotten from various gene sequences.
Variety is another important feature of omics data because they are not restricted to any particular storage format. Speaking of storage format, I am particularly referring to the highly unstructured nature of big data. They can be stored in the form of pictures or images, videos, audio, etc. The penultimate characteristics of big data is its Veracity. Omics data have different level of trustworthiness but one good thing about them is that, because of their very large volume, it is makes up for any inaccuracy. The tendency to have totally inaccurate data is limited since others could make up for the deficits. Veracity is ascribed the most important attribute of big data because it answers the question about trust and data integrity.
lastly, the last V is the data Value. As we earlier explained that data when not analyzed to yield meaningful information and inference is useless. Omics data are highly valuable when they are analyzed by bioinformaticians and statisticians. The information they produce has so far been proven to be of high value in the field of medicine and to be specific, in the management of diseases.
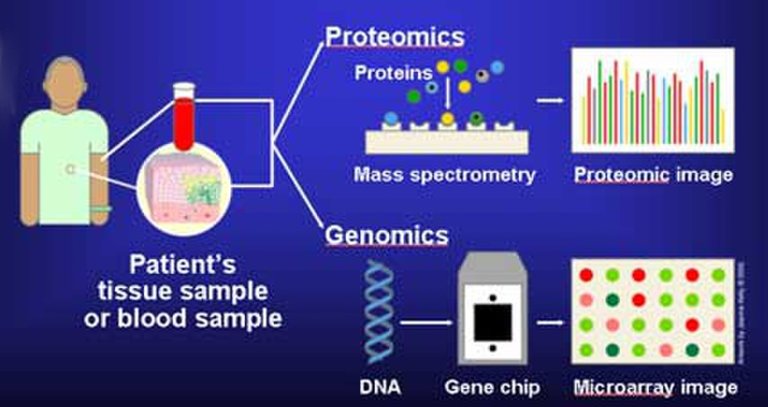
Molecular genetics application in cancer diagnosis
At the gene level (Molecular genetics), omics data are invaluable because they give the clinician the exact information on the aetiology of a disease condition and help narrow drug treatment strategy. This is an area that has so far led to the advancement of precision and personalized medicine.
Molecular biology has drastically changed the landscape of diagnosis because it provides information about a disease at the molecular level. The genes disease causing pathogens like viruses and bacteria can now be sequenced faster than ever. A major advancement has even been observed in cancer treatment.
Before the advent of molecular biology, the regular way of classifying cancer was to examine the supposed tissue after cut up (the process of physically examining a tissue biopsy) through histopathological tissue processing followed by staining and finally microscopy. Under the microscope, depending on the structural morphology and appearance, the cancer is classified.
One of the disadvantage the earlier methodology posed is that, it does not accurately give a clinical indication of the real aetiology of the cancer and this alone can cause wrong treatment approach. Molecular approach to diseases gives a clearer picture of the clinical state of the disease under investigation.
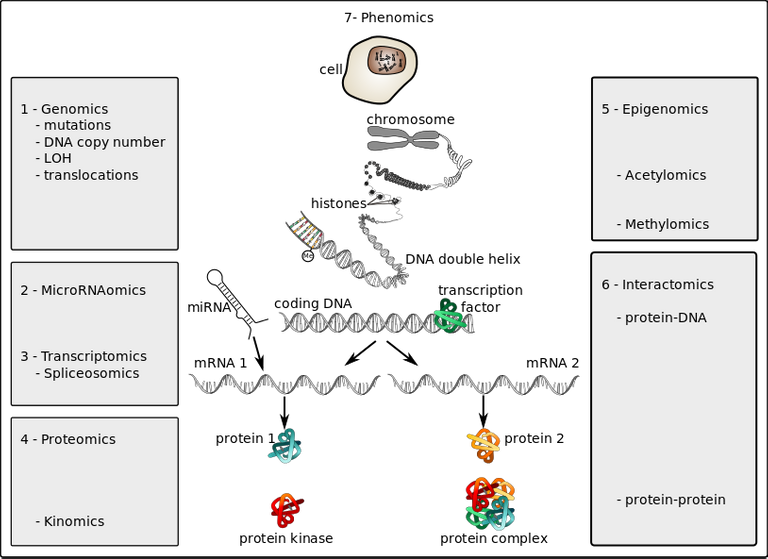
Omics technologies in oncology - cancer
Most times, the omics data can be sources from the protein structure and functions (proteomics), the DNA transcripts (transcriptome), the influence of environmental factors on genes (epigenomics), the physicall expressed traits (phenomics) and how metabolic processed go on in the individual (metabolomics).
How are these omics data sourced from the tissue on a molecular level approach
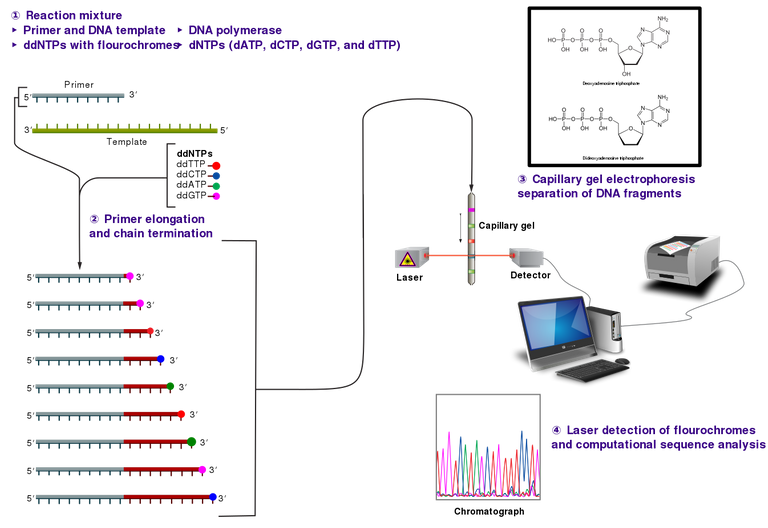
Most of the information extracted and generated from DNA are in the form of codes that are readable through some advanced technologies. Omics data are generated through some technologies and one of the earliest technology to ever be developed was invented by a man named Fredrick Sanger in the year 1977.
His method of DNA (extraction and identification) commonly referred to as the Sanger Sequence used a chain termination DNA that are primed. The method is similar to the conventional polymerase chain reaction. The major difference between his method from the conventional PCR is the incorporation of a modified fluorescent labelled dinucleotides, (dNTPs) called dideoxyribonucleotides, (ddNTPs).
These types of nucleotides do not have one of the most important functional group (3-OH group) for bonding with neighboring nucleotides and as such they break off at random as the polymerase chain reaction continues. For this reason, sanger sequence is also referred to as chain termination sequencing
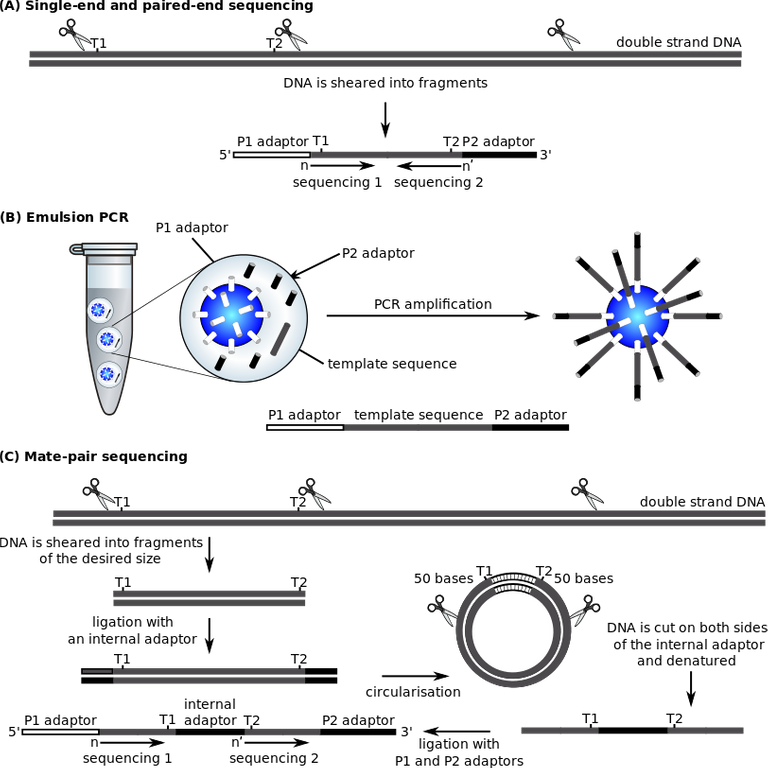
Sanger sequencing is already an old method of gene sequencing and identification because it has been replaced by a more advanced methodology known as the Next Generation Sequencing, NGS. NGS replaced Sanger technique because it offers a high throughput system of constantly generating data and at the same time analyzing these omics data.
NGS has even made it possible to sequence an entire genome of an organism within 24hours, such that was not obtainable with sanger technique. Next generation sequencing is just a name that summarizes all the sequencing techniques currently available (first to fifth generation sequencing techniques).
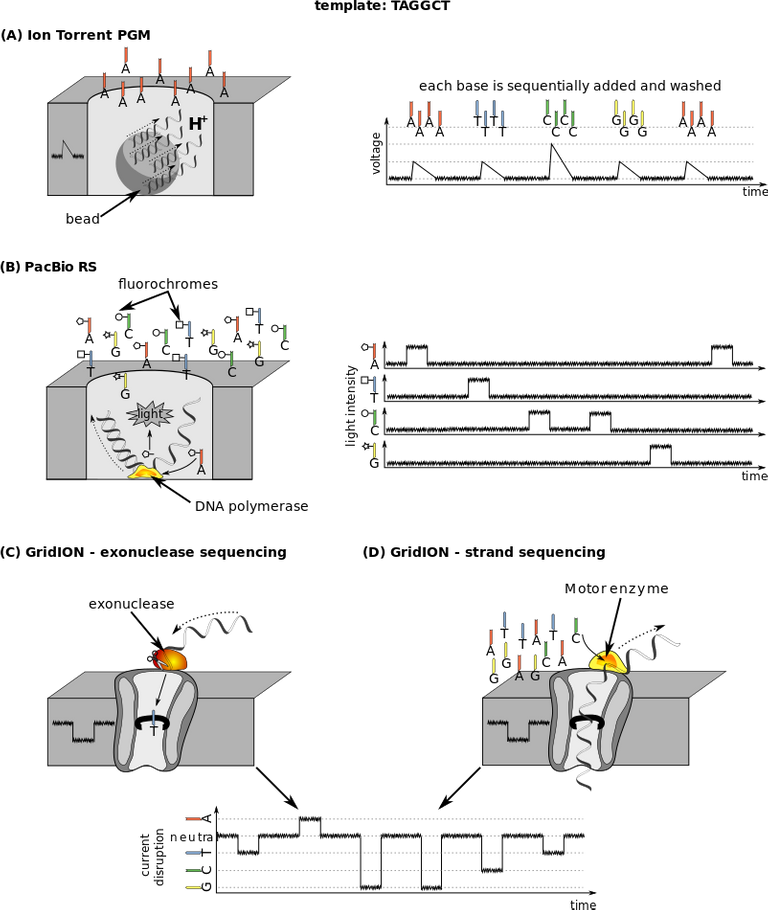
From second to fourth-generation sequencing, illustration on TAGGCT template
All next generation sequencers have four things in common and this include: sample preparation, cluster generation, the actual sequencing of the genes and finally their data analysis for proper interpretation. Pending interpretation, omics data are store in different format like BLAST, SAM/BAM, FASQ and FASTA files. I wont bore you with unnecessary details about this file formats.
The data as we earlier said are mainly store in various repositories for future use by researchers and are later studies systematically by various researchers. Doing this will help in finding out and assessing the epidemiology (study of disease pattern in a population) of a disease condition.
The interpretation of omics data require statistical tools and this is where bioinformatics play huge role. Some of the tools that are available for the analysis of omics data include BLAST, VELVET, EULER, G-MAP and T-bio. These are various (but not limited to) platforms and database through which extracted data sequences can be analyzed and interpreted.
Our future with -Omics data
No doubt genetic data holds a lot of information about diseases and life processes. With the advent of Omics data, we can do phylogenetic studies to trace the history and origin of a disease condition. Omics data can offer us new ways to tackle diseases and also open more avenue to drug discoveries. Today, due to the information gotten from omics data, The Cancer Genome Atlas, TCGA was developed to help in the fight against cancer.
Omics study has made it possible for researchers to study mRNA directly in any tissue sample instead of using cell cultures. Translational projects are now fast on the rise because they offer more benefits than other types of basic research. Since we can sequence the whole genome of an organism or humans, this simply means we can fully understand the genetic bases for virtually all diseases. In essence, it is just a matter of time before the fight against diseases like cancer is successful.
In conclusion, omics studies, offers more potential for the future and anyone who is willing and ready to do exploits in the field of medicine and science in general, see the field of medical biotechnology as an avenue to make impacts. There is more to be discovered. A detailed phylogenetic study on COVID-19 can help us pinpoint the real source of this disease but propaganda and sentiments won't allow this go through. A real blow to researchers I must say. Hope is still alive, someday, it's all gonna be clear and the truth will be unravelled.
Are you confused about your ancestral origin? wake up, its all hidden in your genes. Think Molecular and be sure of finding the real origin through phylogenetic studies.
@cyprianj, bringing molecular genetics at your doorstep on @Stemsocial
Until I come your way again, stay awesome.
References
•Omics-Based Clinical Discovery: Science, Technology, and Applications
•Bioinformatics - a practical guide to the analysis of genes and proteins
•Advances and applications of Bioinformatics in various
fields of life
•The human Genome project
•Sanger sequencing
•Genome
•What Are Omics Sciences?
•Omics technologies in chemical testing