"Citizen Science" Project Activity Report 2 - Simulating top quark production at CERN’s Large Hadron Collider
By Enio...
This article consists of a report of activities in the context of the participatory research project Citizen science particle physics project on Hive led by @lemouth. In it we will see the realization of the second set of activities of the project as described in this article and deals with the simulation of top quark production at CERN's Large Hadron Collider.
Task 1: Top-antitop production at the LHC
I went to the directory where I have the MG5_aMC installation and launched the software. Then I entered the command generate p p p > t t~ which, as we can see in the instructions, serves to define the production of a top-antitop pair in proton-proton collisions.
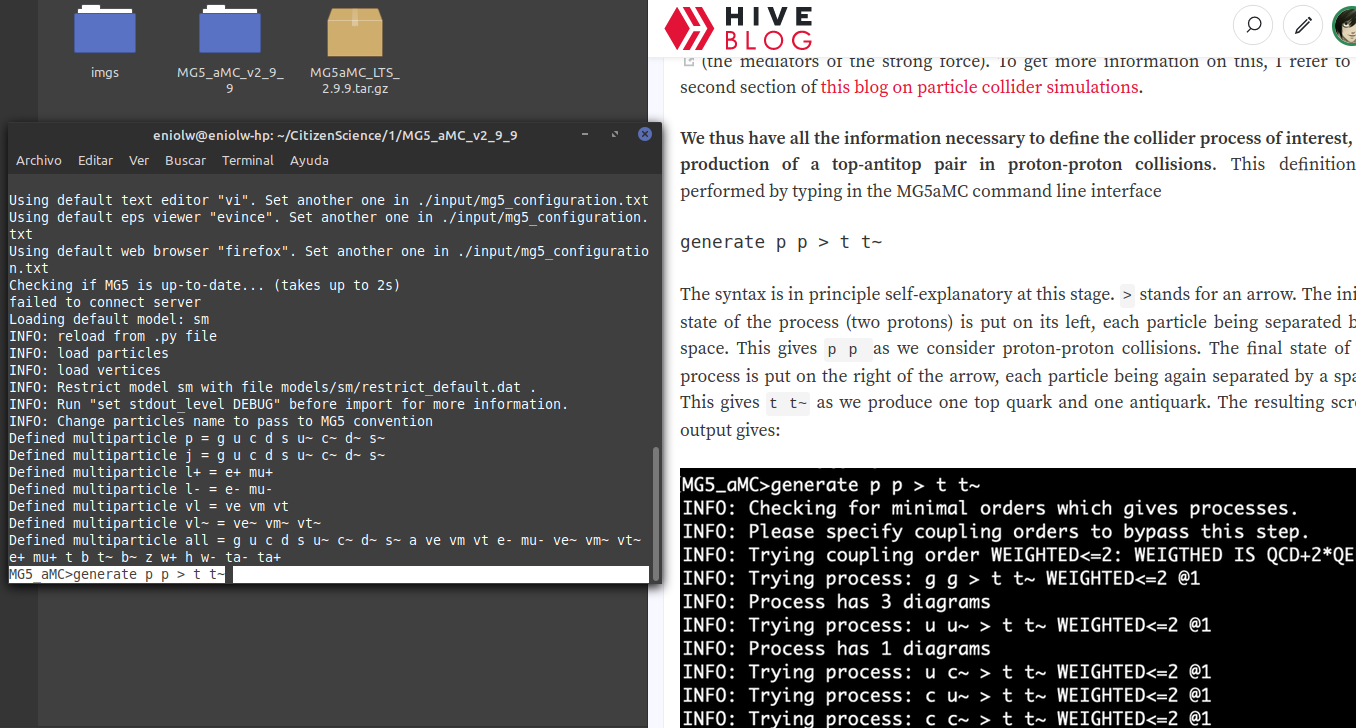
⬆️ Image 1: Entering the first command according to the instructions.
That produced the following output:
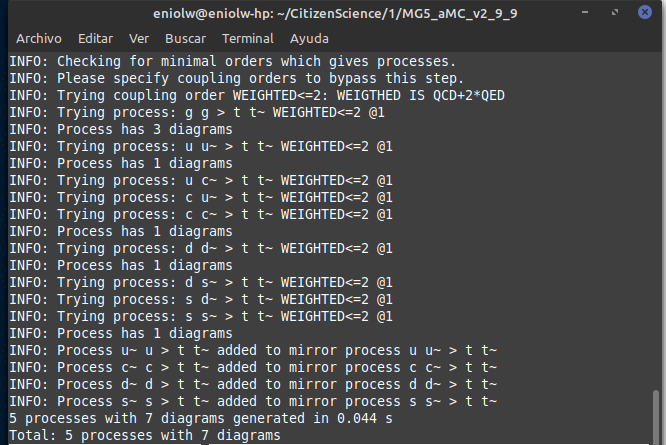
⬆️ Image 2: Everything is normal so far.
I then proceeded to generate the Feynman diagrams with display diagrams, which in turn produced the following output.
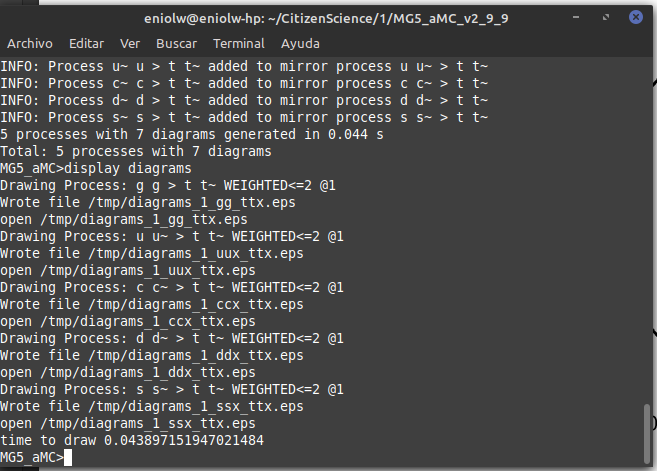
⬆️ Image 3: Second command output.
It says that several .esp files were generated in the /tmp directory. I took a look and indeed they are there. Below is a kind of thumbnail of the 5 files.
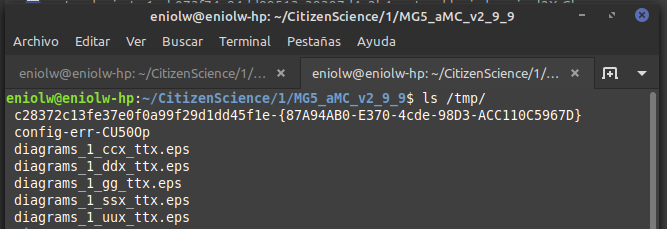
⬆️ Image 4: The /tmp directory showing the just generated files.
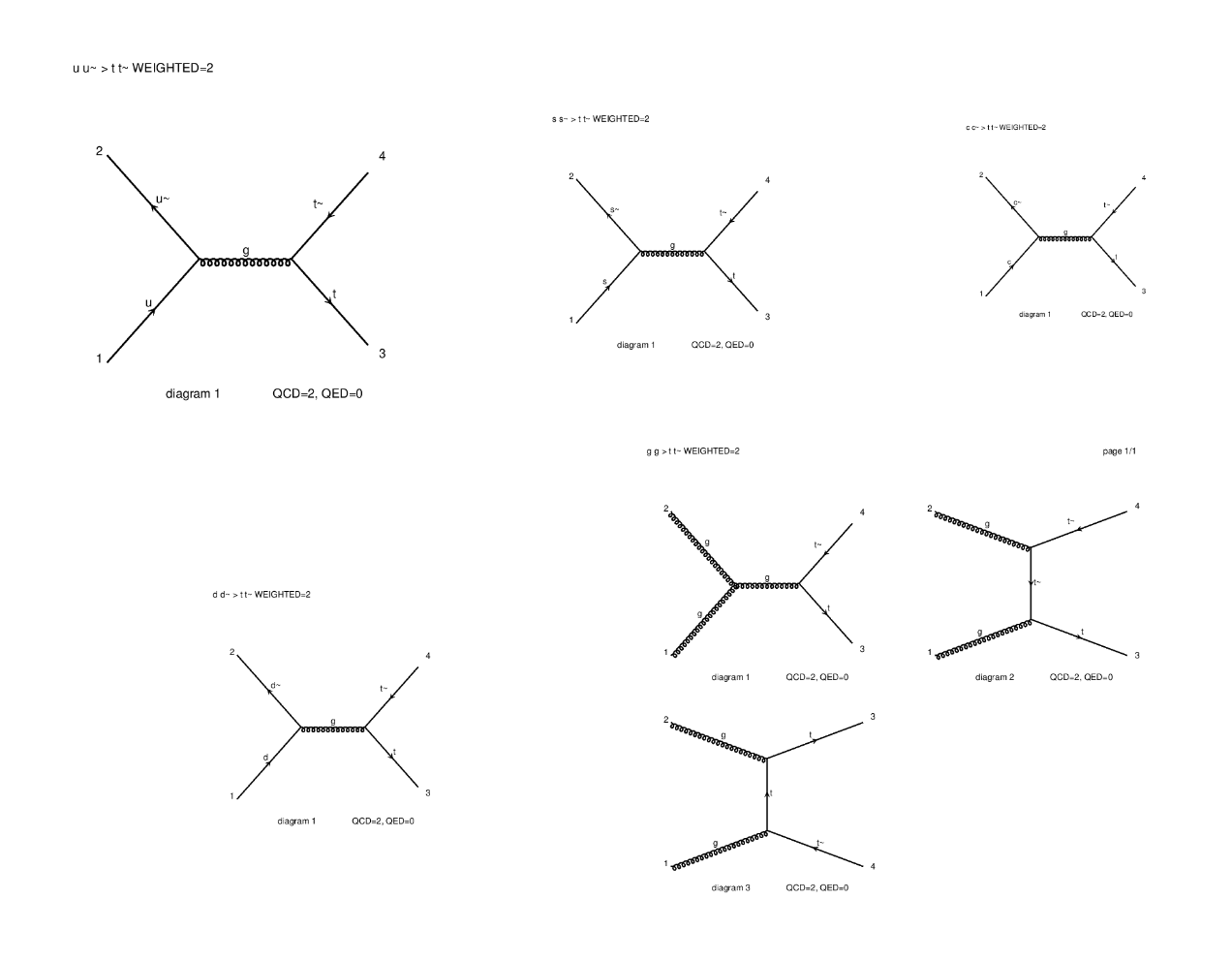
⬆️ Image 5: The five diagrams created (try opening them on another tab to zoom in).
I can't understand these images yet, but according to the instructions we don't need to interpret anything yet.
Task 2: A Fortran code for top-antitop production at the LHC
In order to extract an equation from these diagrams, I have entered the command output fortrancode1. This generates a directory named "fortrancode1" nested in the software directory. Using a new terminal tab I explored the file to verify that the files were generated.
⬆️ Image 6: Third command output.
⬆️ Image 7: Checking the brand new directory "fortrancode1".
Out of curiosity I opened the dgauss.f file in the /source subfolder.
⬆️ Image 8: looking at one of the files out of curiosity.
Task 3: Computing the rate to top-antitop production at the LHC
I went back to MG5aMC to start the process of compiling the Fortran code with launch fortrancode1. This was a bit stressful and in the end I got a compilation error after two attempts. On the first attempt, following the instructions on the interactive menu that appears after using launch, I entered: 1, enter, 4, enter, but failed to edit the line 129 successfully via terminal. The edition process did not behave as expected and I guess it was a problem with Vim. I had to edit using xed, a GUI editor. After that it started running a process for several minutes, but in the end it failed and gave this output:
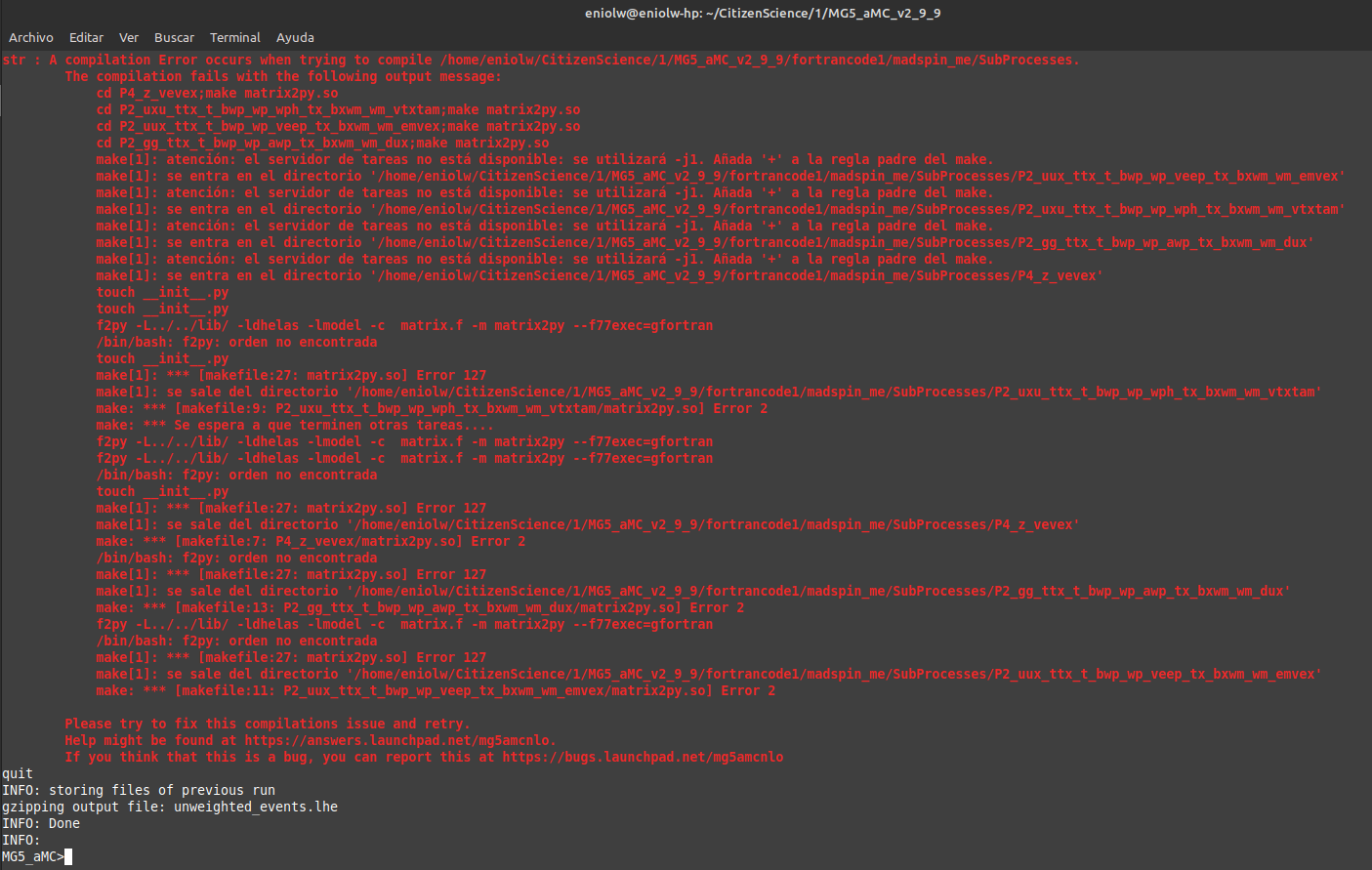
⬆️ Image 9: Something went wrong.
On my second try, I decided to set Nano as the shell text editor to see how it would behave, but in the process I seem to have mistakenly altered another parameter listed in the interactive menu. The truth is that this new compilation also failed. I guess this has something to do with the fact that I changed the line True = use_syst ! Enable systematics studies by False = use_syst ! Enable systematics studies and the tutorial warned that "the code should just crash because some packages are missing…". The point is, I'm not sure if this won't get me through task 4. @lemouth, I appreciate your guidance on this.
In any case, these processes produced an HTML file as output and was automatically opened by the web browser. I attach this view below:
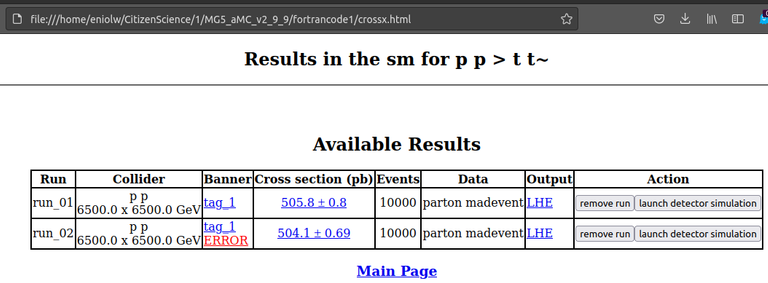
⬆️ Image 10: Simulation output?
This closely resembles the results of the tutorial, so it did work correctly, except for the part I mentioned.
Task 4: Checking out whether everything went fine
When I go to the fortrancode1/Events directory, I don't see the run_01_decayed_1 directory in it, but rather run_01.
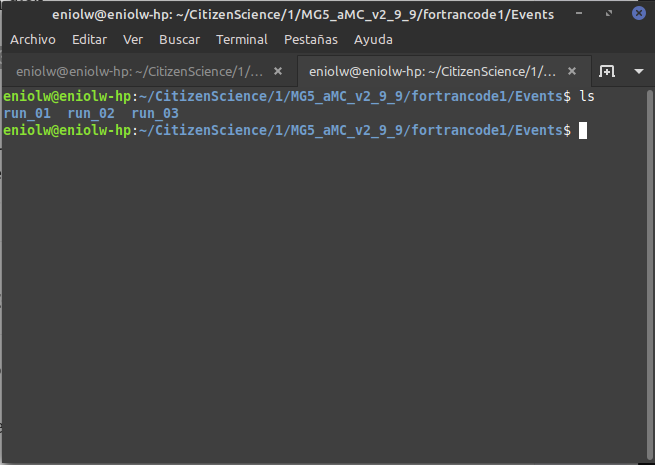
⬆️ Image 11: Actually, it's three directories. I guess I made three attempts after all.
When I go into them and list their contents, I don't see a tag_1_pythia8_events.hepmc.gz file, but this:
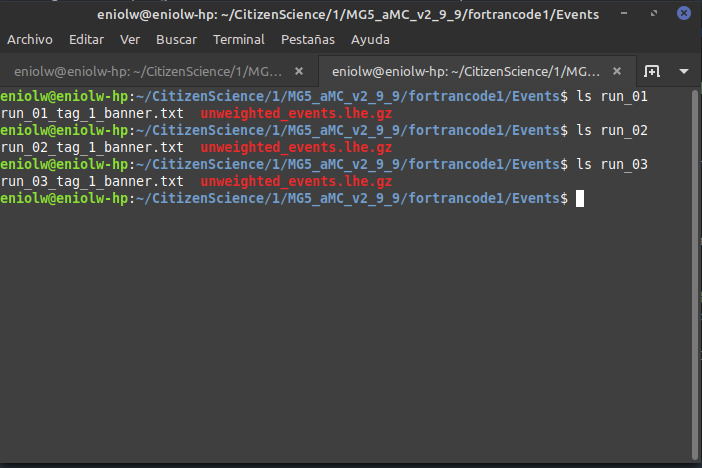
⬆️ Image 12: Something seems missing.
So I don't think I can say that this step is complete :( I hope this information is enough to diagnose the problem. I will be waiting for feedback from @lemouth to correct this and update the report. See you next time.
UPDATE #1 2022/04/05:
After @lemouth's feedback I was finally able to fix the problem by installing NumPy. (Can't believe I didn't have it installed on this machine.) The compilation process took a while and it overheated my computer, but in the end it seems to have worked out well.
⬆️ Image 13: It's working.
However, I couldn't find the Pythia8 output in the log. So I guess it isn't complete yet. I'm pretty sure I enabled Pythia8 by pressing 1 as the instructions say, but I might be wrong, I don't know. What do you think, @lemouth? Should I repeat this? How?
UPDATE #2 2022/04/09:
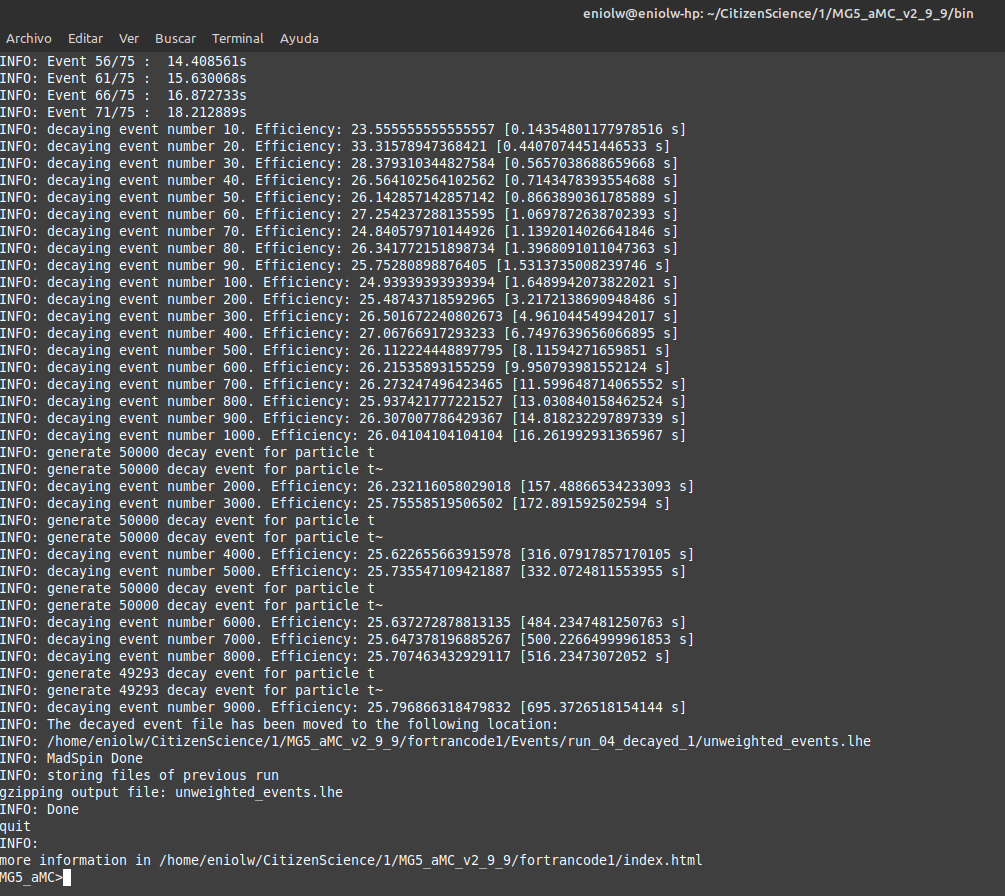
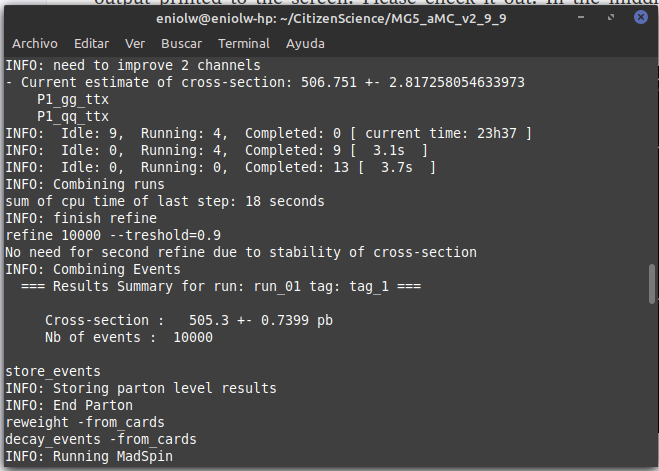
After a few attempts and exchanges with @lemouth, I finally got the expected output. Apparently I messed up a bit my first MG5_aMC installation by changing the name of the root directory where I placed it (I had named it "CivilianScience" instead of "CitizenScience", so I changed it for the second project activity). I noticed it after @lemouth's suggestion to check the mg5_configuration.txt file. I fixed the root folder name in it, but still the program kept crashing. In the end I opted to do a brand new factory installation of MG5_aMC by repeating the procedure of the first activity. With this new installation everything behaved as expected. Below are some screenshots. Thanks, @lemouth, for your help.
⬆️ Image 14: verification output
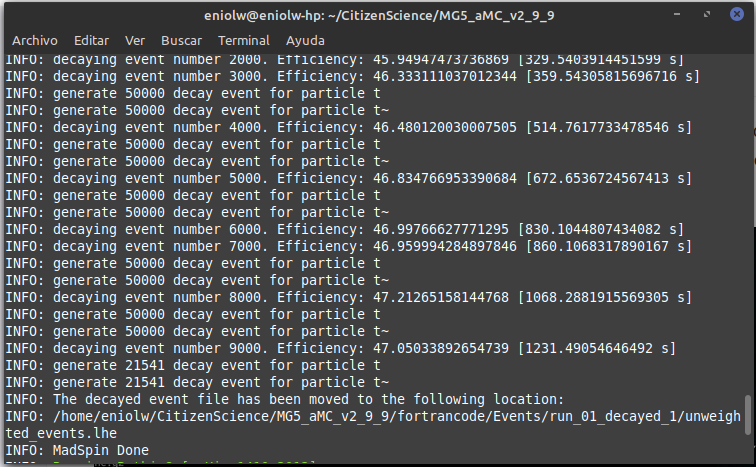
⬆️ Image 15: MadSpin done
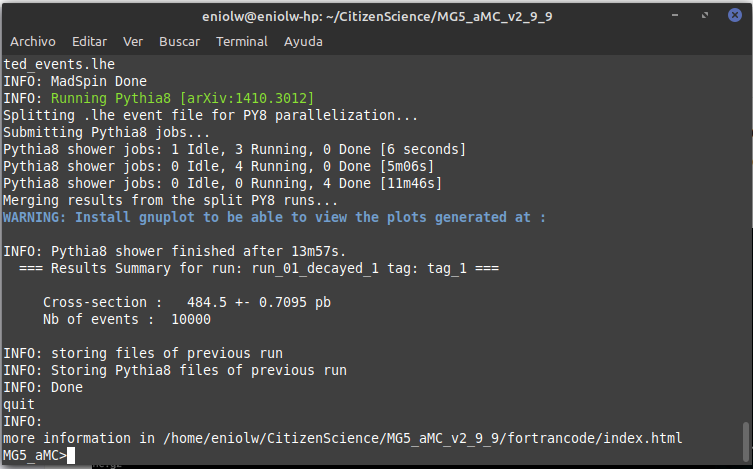
⬆️ Image 16: Pythia8's output
Some References
If you are interested in more STEM (Science, Technology, Engineering and Mathematics) topics, check out the STEMSocial community, where you can find more quality content and also make your contributions. You can join the STEMSocial Discord server to participate even more in our community and check out the weekly distilled.
NOTES
- Unless otherwise stated, the images in this post are the author's.


